Only a small fraction of the human genome (around 2%) contains genes encoding for proteins. The remaining 98% is important for regulation, meaning that it is involved in controlling when and where genes are active. This large portion of the genome produces RNA molecules, called non-coding RNAs, which differ in size, structure and function. As the different types of non-coding RNAs interact with proteins in different ways, big efforts have been put into investigating them. Until now, there were no computational tools available to handle long RNA sequences and studying them through experimental methods is at present a challenge.
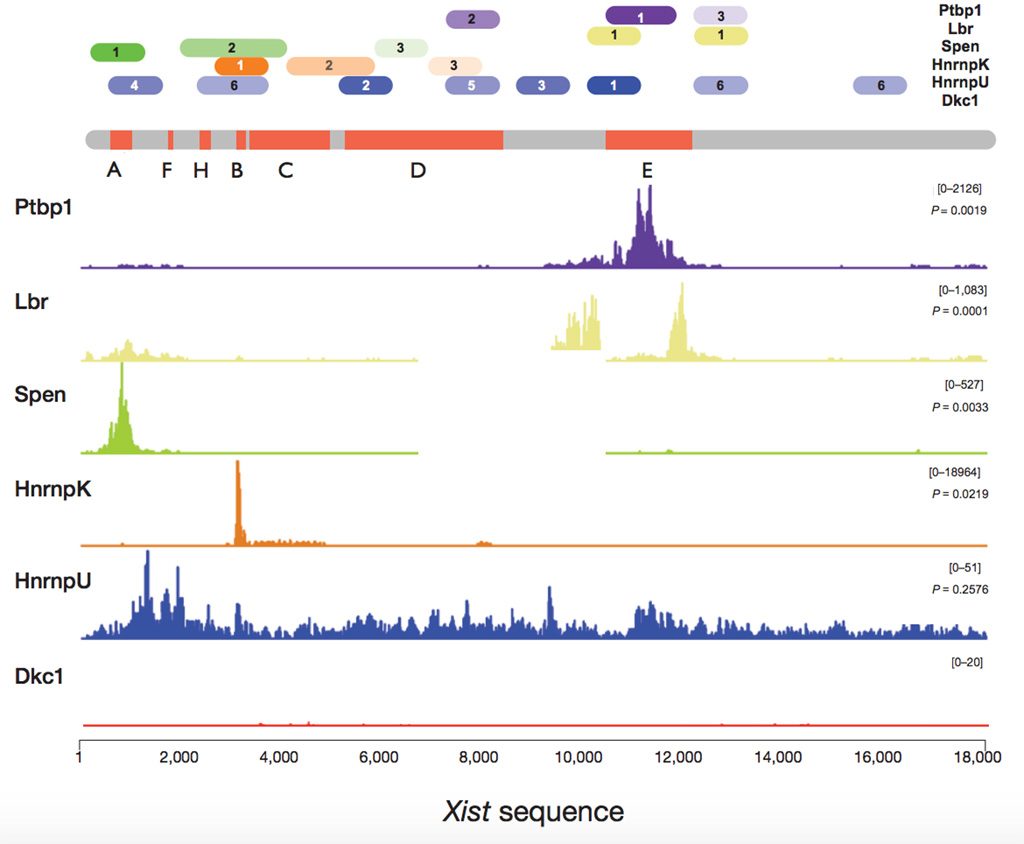
Our new computational tool, catRAPID Global Score, allows us to predict where, along the sequence of a large RNA, a protein will establish a physical contact. To experimentally validate our predictions, we focused on the master regulator of Xchromosome inactivation, Xist. We unveiled the whole protein network interacting with Xist, which is very relevant to understand how X inactivation plays a key role in dosage compensation mechanisms that allow for equal expression of the X and autosomal chromosomes.
Reference