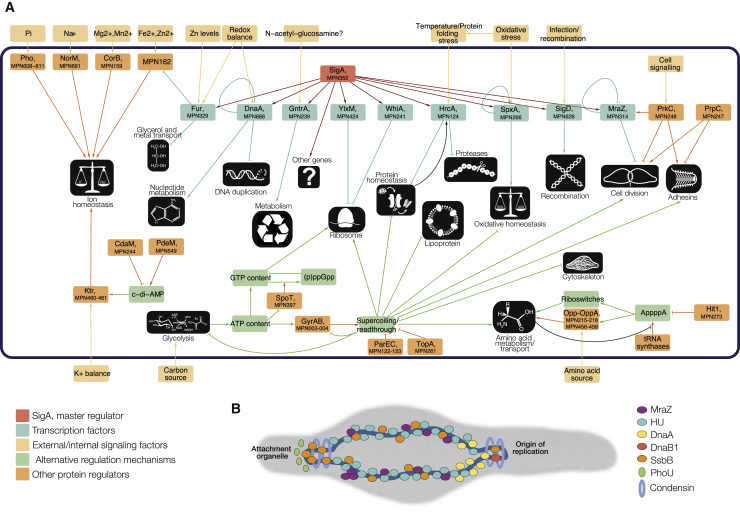
Here, we determined the relative importance of different transcriptional mechanisms in the genomereduced bacterium Mycoplasma pneumoniae, by employing an array of experimental techniques under multiple genetic and environmental perturbations. Of the 143 genes tested (21% of the bacterium’s annotated proteins), only 55% showed an altered phenotype, highlighting the robustness of biological systems. We identified nine transcription factors (TFs) and their targets, representing 43% of the genome, and 16 regulators that indirectly affect transcription. Only 20% of transcriptional regulation is mediated by canonical TFs when responding to perturbations. Using a Random Forest, we quantified the non-redundant contribution of different mechanisms such as supercoiling, metabolic control, RNA degradation, and chromosome topology to transcriptional changes. Model-predicted gene changes correlate well with experimental data in 95% of the tested perturbations, explaining up to 70% of the total variance when also considering noise. This analysis highlights the importance of considering non-TF-mediated regulation when engineering bacteria.
Reference