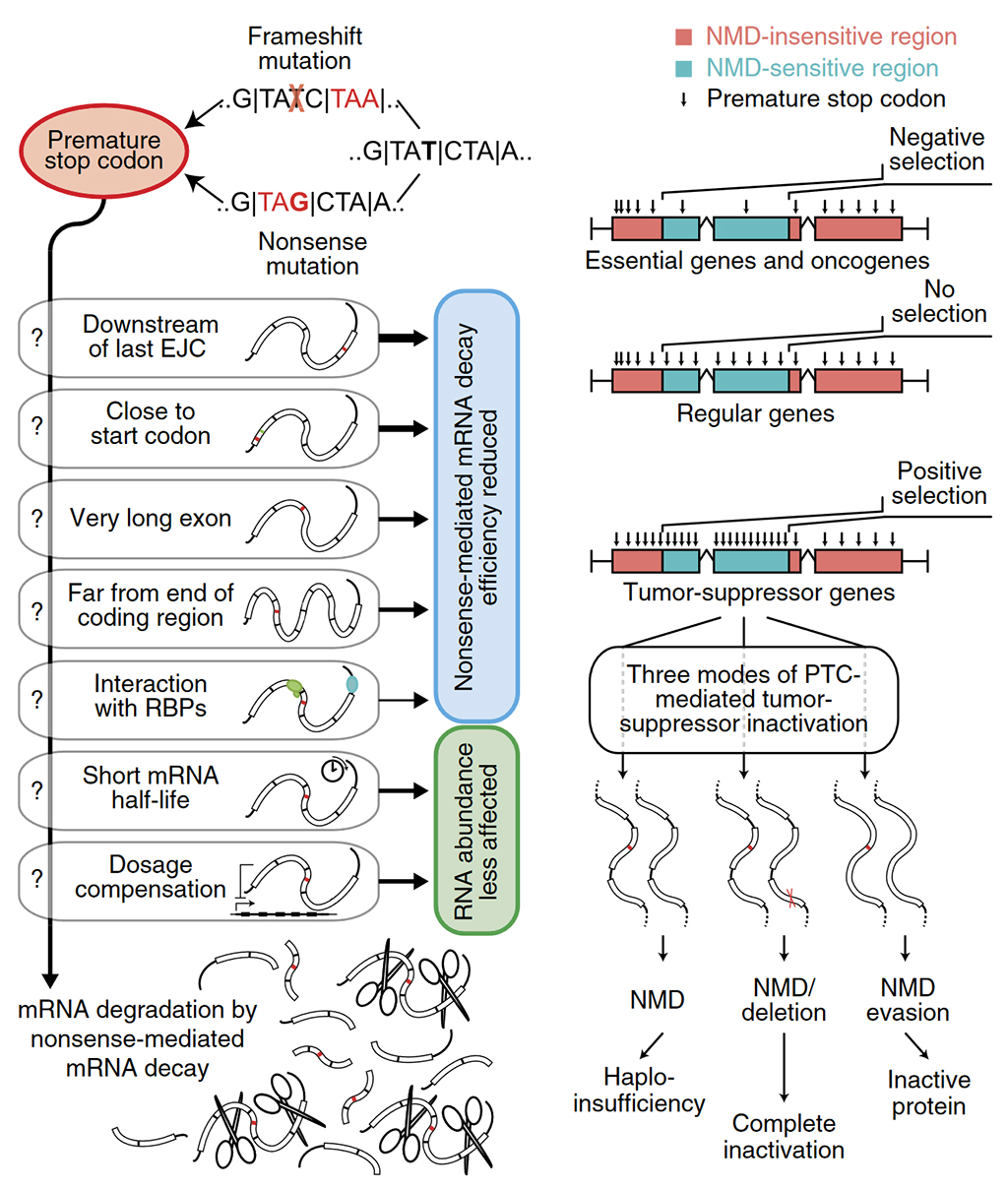
Mutations in DNA can disrupt protein synthesis, sometimes causing truncated proteins which don’t work as intended. Known as nonsense mutations, these types of alterations can give rise to hereditary diseases and different types of cancer. To keep the number of truncated proteins to a minimum, human cells recognise and remove RNAs with nonsense mutations through a quality control process known as nonsense-mediated mRNA decay (NMD).
To better understand the effect of NMD on human disease, we have built NMDetective, a tool describing every possible nonsense mutation that can occur in the human and mouse genomes. Developed by large-scale statistical analyses based on machine learning, the algorithm identifies which mutations in the genome are susceptible to NMD.
As described in Nature Genetics, we used NMDetective to analyse thousands of genetic variants that are known to give rise to hereditary diseases in humans. We were surprised to observe that, in many cases, NMD activity was actually predicted to lead to a greater severity of the disease. This suggests that pharmacological NMD inhibition could slow the progression of many different genetic diseases. To distinguish which patients would benefit from this therapy, it is necessary to apply a precision medicine approach to determine the mutation responsible for the disease and the effect of NMD on this mutation, and this is precisely where NMDetective comes into play.
We also studied the role of NMD in cancer and the interaction between the tumour and the immune system. Remarkably, there is robust genomic evidence that NMD activity is important for the prediction of success of immunotherapy in cancer, because NMD hides mutations that would otherwise trigger the immune system. Therefore, NMDetective can be used to analyse the mutations present in the tumour, in order to better distinguish between cancer patients that respond to immunotherapy from those who do not respond to immunotherapy.
Reference