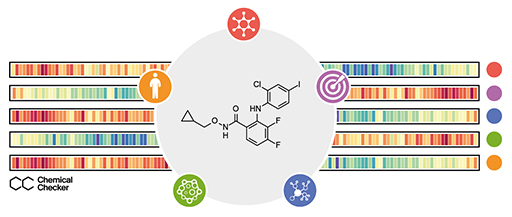
Patrick Aloy – Institut de Recerca Biomèdica (IRB Barcelona)
Small molecules are an excellent tool to probe biological functions and, indeed, they are the main asset of pharmaceutical companies. However, they have received a limited attention by academic researchers during the ‘omics revolution’ since, contrary to gene and protein knowledge, compound data are scattered and diverse, making them inaccessible to most researchers and not suited to standard statistical analyses. Often, the only way to approach the characterization of a compound is to assume it will have the same activity as compounds with similar chemical properties (i.e. the socalled ‘similarity principle’).
The broad release of bioactivity data has led to the realization that the similarity principle applies beyond chemical properties (i.e. molecules eliciting similar side-effects tend to share the mechanism of action, even when their chemical structures appear to be unrelated), suggesting that ‘biological’ similarities offer an alternative means to functionally characterize small molecules. Unfortunately, there is no blueprint to compare the biological profiles of small molecules, since bioactivity data come expressed in formats that are not adapted to common similarity metrics. The Chemical Checker (CC) provides processed, harmonized and integrated bioactivity data on ~1M small molecules. The CC divides data into five levels of increasing complexity, from the chemical properties of compounds to their clinical outcomes. In between, it includes targets, off-targets, networks and cell-level information, such as omics data, growth inhibition and morphology. Bioactivity data are expressed in a vector format, extending the concept of chemical similarity to similarity between bioactivity signatures. We show how CC signatures can aid drug discovery tasks, including target identification and library characterization. We also demonstrate the discovery of compounds that reverse and mimic biological signatures of disease models and genetic perturbations in cases that could not be addressed using chemical information alone. Overall, the CC signatures facilitate the conversion of bioactivity data to a format that is readily amenable for modern machine learning.