Sílvia Osuna – Universitat de Girona (UdG)
Enzyme Design: Enzymes are biomolecules capable of speeding the chemical reactions that take place in our body by many orders of magnitude, making them compatible with life. They are considered the most efficient catalysts known on Earth. Unfortunately, natural enzymes are not well suited for the industrial demands. Enzyme design aims to solve this limitation by introducing changes (i.e. mutations) in the natural enzyme sequence.
One of the most successful strategies for designing new enzymes for industrially-relevant reactions and conditions is experimental laboratory evolution. Although highly powerful, it has also a high economical cost associated, which hampers the extensive application of enzymes in industry. Computational enzyme design has the potential of predicting the set of specific changes required for novel enzymatic function, but so far none of the available strategies has been able to provide new enzymes with efficiencies rivaling those of natural and laboratory engineered enzymes.
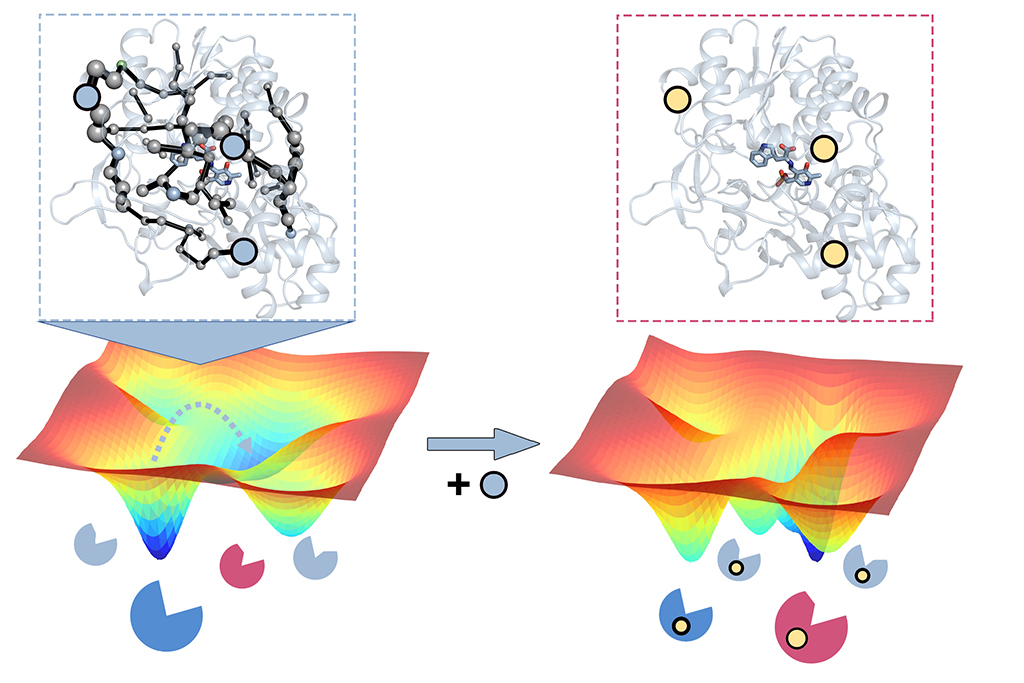
Predicting distal functional mutations: The mutations introduced in many laboratory evolution experiments are often located all around the enzyme structure, which contrasts with computational enzyme design that reduces the problem into alterations in the region where the reaction happens (also called the active site).
How can we rationally predict distal activity-enhancing mutations? Given the large number of possible positions to mutate in natural enzymes, the computational prediction of such distal mutations impacting function has been proven to be extremely challenging. We have developed new computational tools that allow, for the first time, the prediction of mutations in the enzyme active site, but also at positions located distal from where the reaction occurs. This has been achieved by accurately considering the enzyme ability to adopt multiple conformations key for their enzymatic function.